Capricorn
Capricorn

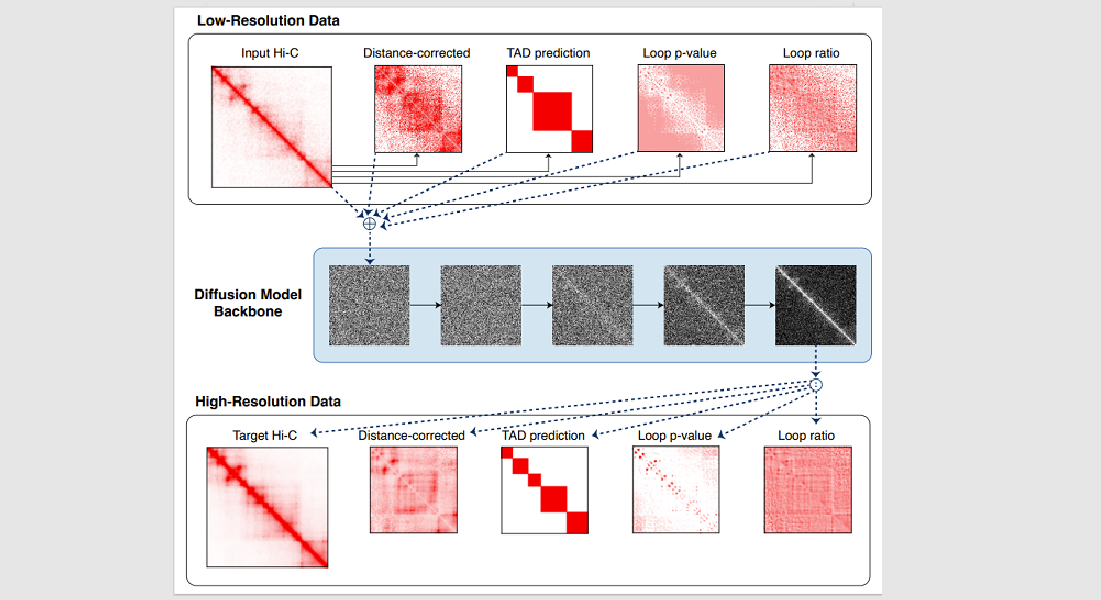
高分辨率的Hi-C接触矩阵测量基因组的详细三维结构,但高分辨率的实验Hi-C数据生成成本高昂,而且相对罕见。
目前有一些增强低分辨率接触矩阵的计算方法,但主要基于自然图像的分辨率增强方法,因此经常使用不区分生物意义接触和背景接触的模型。
我们介绍了Capricorn,这是一种用于Hi-C分辨率增强的工具,它将高阶染色质特征作为输入Hi-C接触矩阵的附加视图,并利用扩散概率模型主干来生成高分辨率矩阵。
我们表明,Capricorn在跨细胞系设置中优于最先进的方法,对从生成的高分辨率数据调用的循环,其均方误差降低了17.8%,F1分数提高了22.9%。
我们还表明,Capricorn在交叉染色体设置中表现良好,与现有方法相比,下游Loop-F1得分再次提高15.7%。论文:Enhancing Hi-C contact matrices for
loop detection with Capricorn, a multi-view diffusion model。 Capricorn的实现和源代码在https://github.com/CHNFTQ/Capricorn。本论文被ISMB-2024会议接收并将发表在Bioinfomatics期刊上。
High-resolution Hi-C contact matrices measure the detailed three-dimensional architecture of the
genome, but high-resolution experimental Hi-C data are expensive to generate and relatively rare. Computational
methods
to enhance low-resolution contact matrices exist but are largely based on resolution
enhancement methods for natural images and hence often employ models that do not distinguish between biologically
meaningful contacts and background contacts. We present Capricorn, a tool for Hi-C
resolution enhancement that incorporates high-order chromatin features as additional views of the input
Hi-C contact matrix and leverages a diffusion probability model backbone to generate a high-resolution
matrix. We show that Capricorn outperforms the state-of-the-art in a cross-cell-line setting, improving existing
methods by 17.8% in mean-squared error and 22.9% in F1 score for loops called from the
generated high-resolution data. We also show that Capricorn performs well in the cross-chromosome setting, again
improving the downstream loop F1 score by 15.7% relative to existing methods. Paper: Enhancing Hi-C contact matrices for
loop detection with Capricorn, a multi-view diffusion model. Capricorn's
implementation and source code are available at https://github.com/CHNFTQ/Capricorn.This paper is accepted by ISMB-2024 and to be published on Bioinfomatics journal.